背景:近期主要是在ROC曲线上的问题,数据集是titanic、heart disease 以及python自带的数据集iris
参考链接1:混淆矩阵(https://blog.csdn.net/seagal890/article/details/105059498)
参考链接2:如何画ROC曲线
参考链接3:Sklearn.metrics评估方法
参考链接4:用Python绘制ROC曲线
混淆矩阵(confusion matrix)
基本上所有的模型评价指标最后都是从cm这一块延伸出来的。
模型评价指标
- 精确率(accuracy)、F-score是针对总体值来说的。
- Precision、Recall、Specificity等主要针对每一行来说
- ROC曲线:对于ROC,最重要的三个概念就是TPR、FPR和截断点()
举例分析
- 1.iris数据集
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm,datasets
from sklearn.metrics import roc_curve,auc
from sklearn import model_selection
iris = datasets.load_iris()
X = iris.data
y = iris.target #都是array数据,看着不舒服
#转化无dataframe数据
X_df = pd.DataFrame(X)
y_df = pd.DataFrame(y)
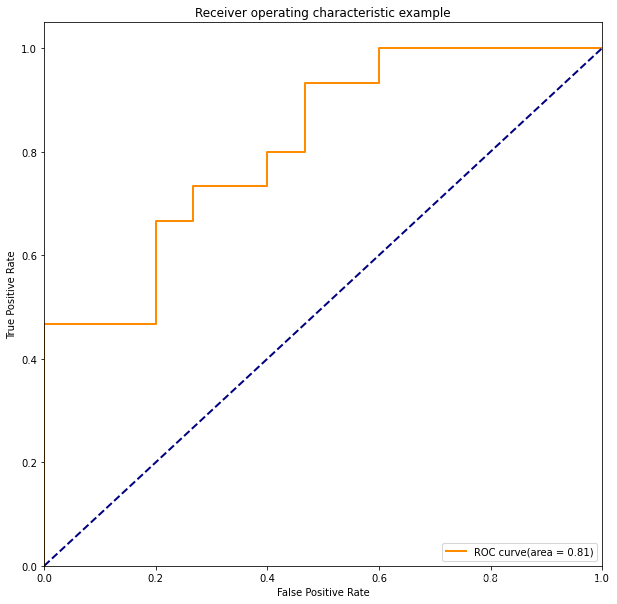
效果图

最出色的部分在于计算分类值
y_df.value_counts()
0 50
1 50
2 50
dtype: int64
X,y = X[y!=2],y[y!=2] #转化为二分类问题,数据减少
X.shape #原本是150个数据,减少为属于0,1类的数据了
(100, 4)
#训练集和测试集的划分
X_train,X_test,y_train,y_test = model_selection.train_test_split(X,y,test_size=0.3,random_state=0)
#你看X分为2份,y分为两份
X_train.shape
(70, 804)
y_train.shape
(70,)
#支持向量机SVM
svm = svm.SVC(kernel="linear",probability=True,random_state=random_state)
y_score = svm.fit(X_train,y_train).decision_function(X_test)
y_score.shape
(30,)
##计算真正率和假正率
fpr,tpr,threshold = roc_curve(y_test,y_score)
roc_auc = auc(fpr,tpr)
fpr.shape
(14,)
#绘图ROC
plt.figure()
lw = 2
plt.figure(figsize=(10,10))
plt.plot(fpr,tpr,color="darkorange",
lw = lw,label = "ROC curve(area = %0.2f)"%roc_auc)
plt.plot([0,1],[0,1],color="navy",lw=lw ,linestyle = '--')
plt.xlim([0.0,1.0])
plt.ylim([0.0,1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc = "lower right")
plt.show()
- 2.heart disease数据集
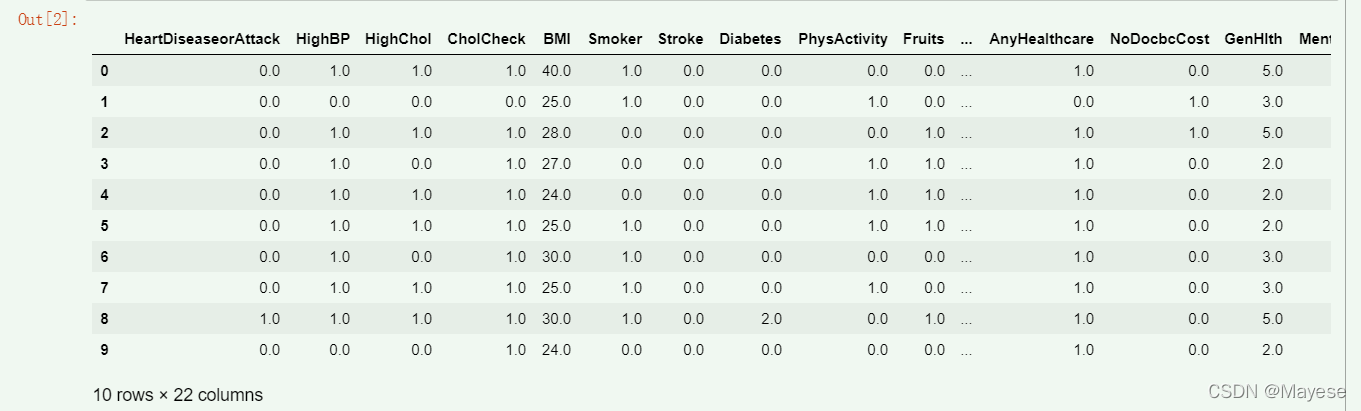
该数据集最大的优点在于没有缺失值
dataset = pd.read_csv("heart disease.csv")
dataset.head(10)
dataset.isnull().sum() #很好不存在缺失值,数据预处理也不做了,直接进行建模
data = dataset.rename(columns={"HeartDiseaseorAttack":"target"})
#导入绘图工具包
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
#错误提醒
import warnings
warnings.filterwarnings("ignore")
#show the counts of observation in each categorical bin using bars
sns.countplot(data['target']) #对于标签类,患病还是不患病
print(data.target.value_counts())
data["target"] = [1 if i == 1.0 else 0 for i in data.target] #转化为整形int
y = data.target
x = data.drop(['target'],axis = 1)
columns = x.columns.tolist()
columns
['HighBP',
'HighChol',
'CholCheck',
'BMI',
'Smoker',
'Stroke',
'Diabetes',
'PhysActivity',
'Fruits',
'Veggies',
'HvyAlcoholConsump',
'AnyHealthcare',
'NoDocbcCost',
'GenHlth',
'MentHlth',
'PhysHlth',
'DiffWalk',
'Sex',
'Age',
'Education',
'Income']
#for machine learning
from sklearn.preprocessing import StandardScaler,MinMaxScaler #标准化
from sklearn.model_selection import train_test_split,GridSearchCV #训练集和测试集的划分 GridSearcherCV没有用到呀
from sklearn.neighbors import KNeighborsClassifier,NeighborhoodComponentsAnalysis
from sklearn.decomposition import PCA #decomposition指分解
from sklearn.metrics import confusion_matrix, f1_score, precision_score, recall_score
from sklearn.model_selection import KFold, cross_val_score, train_test_split
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
from sklearn.svm import SVC
# from sklearn.preprocessing import StandardScaler
import seaborn as sns
#train test split
X_train,X_test,Y_train,Y_test = train_test_split(x,y,test_size=0.3,random_state=42) #按照0.7 和0.3的比例来划分
#标准化
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
X_train_df = pd.DataFrame(X_train,columns= columns) #生成数据表
X_train_df_describe = X_train_df.describe()
X_train_df['target']= Y_train
#knnscore = knn_param_search.score(X_test,Y_test) 这个如果不使用呢?
knncm = confusion_matrix(Y_test,y_pred) #混淆矩阵:Y_test,y_pred
#knncr = classification_report(Y_test,y_pred)
acc = metrics.accuracy_score(Y_test,y_pred) #Y_test,y_pred
prec = metrics.precision_score(Y_test,y_pred)
rec = metrics.recall_score(Y_test,y_pred)
f1 = metrics.f1_score(Y_test,y_pred)
cm = confusion_matrix(Y_test,y_pred)
Specificity = cm[0,0]/(cm[0,0]+cm[0,1])
Sensitivity = cm[1,1]/(cm[1,0]+cm[1,1])
print('K Nearest Neighbour')
print('*******************')
print('Testscore')
print('---------')
#print(knnscore)
print('\n')
print('confusion Matrix')
print('----------------')
print(cm)
print('\n')
print('Classification Report')
print('---------------------')
#print(knncr)
print('Accuracy')
print('---------------------')
print(acc)
print('Precision')
print('---------------------')
print(prec)
print('Recall')
print('---------------------')
print(rec)
print('F1_score')
print('---------------------')
print(f1)
print('Specificity')
print('---------------------')
print(Specificity )
print('Sensitivity ')
print('---------------------')
print(Sensitivity )
# Compute False postive rate, and True positive rate
fpr, tpr, thresholds = metrics.roc_curve(Y_test,y_pred) #所以必须使用网格参数搜索了
# Calculate Area under the curve to display on the plot
roc_auc = metrics.roc_auc_score(Y_test,y_pred)
#fpr, tpr, thresholds = metrics.roc_curve(y_test,y_probs)
#roc_auc = auc(fpr, tpr) #auc为Roc曲线下的面积
plt.title('Receiver Operating Characteristic of KNN')
plt.plot(fpr, tpr, 'b', label = 'AUC = %0.2f' % roc_auc)
plt.legend(loc = 'lower right')
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0, 1])
plt.ylim([0, 1])
plt.ylabel('True Positive Rate')
plt.xlabel('False Positive Rate')
plt.show()
print("ROC_AUC of KNN = ",roc_auc)
版权声明:本文为weixin_45081871原创文章,遵循 CC 4.0 BY-SA 版权协议,转载请附上原文出处链接和本声明。